Taxonomic trees are ubiquitous in biodiversity software. A very common application is using a tree to allow the users to browse the data. Other applications are: training classification models, curating a collection, visualizing research results etc.
Data is often stored in a relational database, such as MySQL. Unfortunately, relational databases are not particularly well suited for storing tree structures. Yet the choice of a database may be guided by more important requirements, and so the taxonomic tree is sometimes implemented as an afterthought. The result can be a structure that is difficult to maintain and to query, sometimes requiring more work than expected to maintain and finally yielding a less satisfying experience for the end user.
I will show some counterexamples, and how get a better result by using a data structure called “nested set”.
Don’t do this
The flat table
Data is often collected in tabular format. So if you have – for example – a list of occurrences, chances are you have a row for each occurrence, with a column for the corresponding species. It is tempting to replicate this flat table structure to store taxonomic information in the database, adding a column for genus, family, order and class. The result is a flat table with a line for each species (or technically each OTU) and a column for higher taxonomic ranks. While this is easy to grasp, it also has major drawbacks:
- To make the data searchable, you’ll have to index all the columns, which in turn makes adding data a lot slower, as indexes have to be rebuilt for each insert operation.
- When an update of the taxonomy is necessary (which happens surprisingly often), a complex subset of the data has to be edited, which is error-prone.
A table for each rank
Another tempting pattern is to make a table for each taxonomic rank, so one table for species, one for genus, family, order, class as required. This pattern takes care of the indexing problem caused by flat tables, but actually makes things worse:
Querying such a structure requires joins between 5 or more tables, which can be complex and error-prone.
The linked list
You could have a flat table with a column for child-to-parent relationships. This has the advantage that all the data is in one table, however traversing a linked list is not a stroll in the park:
Traversing a linked list often involves recursive algorithms, which are at the very least hard to wrap your head around.
Clarifying the assumptions
In order to find a satisfying solution, let’s take a step back. Taxonomy actually pre-dates the theory of evolution by more than a century. Linnaeus published his Systema Naturae in 1735, while Darwin’s On the Origin of Species is from 1859. Therefore taxonomy is primarily a system for classification, the – albeit essential – assumption that all lifeforms stem from a single lineage was added afterward.
Therefore, a “tree of life” can be be interpreted in at least two different ways: in a linaean sense, it’s a classificatin system, where each node of the tree represents a classification rank (order, class etc.). In a darwinian sense, each node of the tree stands for the most recent common ancestor of a lineage.
Whichever interpretation you are interested in, a tree can be described mathematically using a data structure called a “nested set”. A set being a list of unique items, with no empty values, and nested meaning that these items can themselves be sets.
Nested sets
So the problem of implementing a taxonomic tree in a relational database is equivalent to building a structure that can hold a nested set. There are several standard solutions for that, but a common and well tested one employs “right” and “left” indexes. This is illustrated in figure 1.
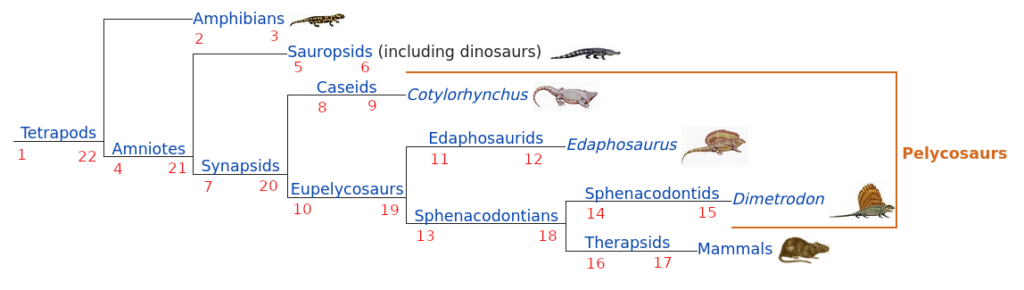
Figure 1 shows a taxonomic tree (“The ancestry of mammals”, from Wikipedia) where I added right and left indexes. To compute the indexes, start at the left of the image, increase by one on each branch and turn around once you get to a leaf. It’s quite intuitive after you get a grip on it, and writing the algorithm is very easy.
Once the indexes are added to each taxon, navigating the tree is simple. For example:
Which taxa are part of the group Synapsids?
All entries with a left index lft so that: 7 < lft < 20
Which group are Therapsids part of?
Take the left and the indexes of Therapsids (lft and rgt) and find the group with the largest left index lfx larger than lft and the smallest right rgx index larger than rgt: max( lfx < lft ) & min(rgx > rgt)
What about Pelycosaurs?
Now it becomes interesting: Pelycosaurs are a group that is used in older publications, but is presently considered deprecated. This kind of taxonomy change happens quite often. Don’t panic, Pelycosaurs are the group between left index 8 and right index 15, therefore they are Synapsids, but do not include mammals.
Implementation
Below is a basic implementation of a taxonomic tree as nested set in a relational database table. Additionally to the left and right indexes (lft and rgt), I added a unique identifier pulled from the Open Tree of Life (more on that in the next post) as well as the scientific name and vernacular names in two languages.
Note the “rank” column, you’re going to need that to filter by taxon rank (e.g. by species) when querying the tree.
Only the left and right indexes columns have to be… indexed. That’s all that’s required to find your way around the tree, no joins, no recursion.
There’s however a trade-off: each time you add a new species, you’ll have to re-compute some of the left and right indexes. However, it’s just a counting operation, no complex queries involved.
CREATE TABLE IF NOT EXISTS `taxon` (
`ott_id` INT NOT NULL COMMENT 'The Open Tree of Life id of this taxon.',
`lft` INT NULL,
`rgt` INT NULL,
`unique_name` VARCHAR(1024) NOT NULL COMMENT 'The scientific name of this taxon.',
`rank` ENUM('subspecies', 'species', 'genus', 'family', 'order', 'class', 'phylum') NOT NULL COMMENT 'The rank of this taxon.',
`vernacular_name_english` VARCHAR(1024) NULL,
`vernacular_name_german` VARCHAR(1024) NULL,
PRIMARY KEY (`ott_id`),
INDEX `taxon_lftx` (`lft` ASC),
INDEX `taxon_rgtx` (`rgt` ASC),
ENGINE = InnoDB;
In a next post, I’ll show how to populate a tree by pulling data from the Open Tree of Life.